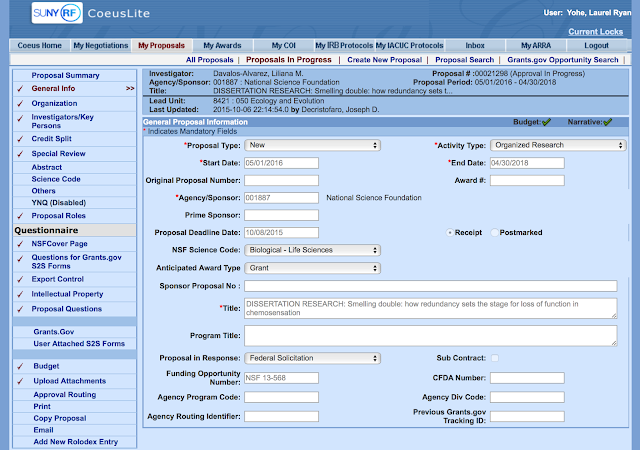
We are looking for relaxed selection in Trpc2 of several lineages of bats that have reduced morphology of vomeronasal system organs or brain regions.
To run RELAX:
RELAX is relatively recent and you have to download the most recent batch files for HyPhy.
Start HyPhy.
Open->Open Batch File...
Navigate to Applications/hyphy/res/TemplateBatchFiles/
Select RELAX.bf
It will automatically prompt you to select your data/alignment file.
Then select your tree file.
Make sure the correct number of Ts, Rs, and Us match.
It should hopefully begin, assuming you have your tree set up correctly and your alignment does not have any stop codons AND it is in frame.
Something VERY critical if you are running it in the GUI...you must select the test branches directly when it asks for it, not just when you put them in the tree. Otherwise it estimates the reference branches as the test branches and vice versa.
RELAX1:
Alignment: bat_data_match.nex
Tree: bat_tree_match_RELAX1.tre
(((((((((((((((((((((Artibeus_jamaicensis{R}:2.431373248,Artibeus_obscurus{R}:2.431373248){R}:0.3588412223,Artibeus_fimbriatus{R}:2.79021447){R}:0.9561230126,(Artibeus_intermedius{R}:0.6313580609,Artibeus_lituratus{R}:0.6313580609){R}:3.114979422){R}:1.424889238,Artibeus_inopinatus{R}:5.171226721){R}:0.9218212476,Artibeus_concolor{R}:6.093047969){R}:2.392412914,((((Artibeus_watsoni{R}:2.760434077,Artibeus_incomitatus{R}:2.760434077){R}:1.940812275,Artibeus_phaeotis{R}:4.701246352){R}:0.9218549563,Artibeus_cinereus{R}:5.623101308){R}:0.8637586984,(Artibeus_glaucus{R}:3.316302056,Artibeus_gnomus{R}:3.316302056){R}:3.170557951){R}:1.998600876){R}:2.413693209,(((Ariteus_flavescens{R}:2.487096883,Ardops_nichollsi{R}:2.487096883){R}:2.828400699,Phyllops_falcatus{R}:5.315497583){R}:0.8401618707,((Pygoderma_bilabiatum{R}:3.602361855,Sphaeronycteris_toxophyllum{R}:3.602361855){R}:1.181679598,Centurio_senex{R}:4.784041453){R}:1.371618001){R}:4.743494639){R}:1.13714402,Ectophylla_alba{R}:12.03629811){R}:0.9420299332,Enchisthenes_hartii{R}:12.97832805){R}:1.825921019,((((((((Platyrrhinus_nigellus{R}:2.504001415,Platyrrhinus_dorsalis{R}:2.504001415){R}:0.4677903585,(Platyrrhinus_infuscus{R}:2.110403375,Platyrrhinus_aurarius{R}:2.110403375){R}:0.8613883988){R}:1.095202405,Platyrrhinus_albericoi{R}:4.066994179){R}:2.177118584,Platyrrhinus_lineatus{R}:6.244112763){R}:1.779671702,Vampyrodes_caraccioli{R}:8.023784464){R}:2.695962001,(Vampyressa_thyone{R}:7.747059999,Mesophylla_macconnelli{R}:7.747059999){R}:2.972686466){R}:0.777302653,(((Chiroderma_villosum{R}:1.968379874,Chiroderma_improvisum{R}:1.968379874){R}:1.432916649,Chiroderma_doriae{R}:3.401296523){R}:5.790148924,(Vampyressa_brocki{R}:7.444560681,Vampyressa_bidens{R}:7.444560681){R}:1.746884766){R}:2.305603671){R}:0.737544275,Uroderma_magnirostrum{R}:12.23459339){R}:2.569655671){R}:2.978950491,(((Sturnira_bogotensis{R}:4.351270017,Sturnira_tildae{R}:4.351270017){R}:0.6450817233,Sturnira_magna{R}:4.99635174){R}:0.4749192885,(Sturnira_oporaphilum{R}:0.9163878452,Sturnira_lilium{R}:0.9163878452){R}:4.554883183){R}:12.31192853){R}:1.896666128,Rhinophylla_fischerae{R}:19.67986568){R}:1.622496147,(((((Carollia_perspicillata{R}:2.166884003,Carollia_brevicauda{R}:2.166884003){R}:0.7022808564,Carollia_sowelli{R}:2.869164859){R}:1.049593577,Carollia_subrufa{R}:3.918758436){R}:4.394363478,Carollia_castanea{R}:8.313121914){R}:11.76400773,(Glyphonycteris_sylvestris{R}:15.43079994,Trinycteris_nicefori{R}:15.43079994){R}:4.6463297){R}:1.225232188){R}:1.57140971,((((Lonchophylla_hesperia{R}:9.371624678,Lionycteris_spurrelli{R}:9.371624678){R}:2.706833404,Platalina_genovensium{R}:12.07845808){R}:1.204240126,Lonchophylla_chocoana{R}:13.28269821){R}:2.390290187,Lonchophylla_mordax{R}:15.6729884){R}:7.200783145){R}:1.155625037,((((((Glossophaga_longirostris{R}:5.673316434,Glossophaga_commissarisi{R}:5.673316434){R}:1.473493579,Glossophaga_soricina{R}:7.146810013){R}:4.41746123,(Leptonycteris_nivalis{R}:1.825719598,Leptonycteris_curasoae{R}:1.825719598){R}:9.738551645){R}:6.408254338,(Brachyphylla_pumila{T}:1.377027909,Brachyphylla_cavernarum{T}:1.377027909){T}:16.59549767){R}:1.44044124,(((Musonycteris_harrisoni{T}:1.250127196,Choeronycteris_mexicana{T}:1.250127196){T}:4.862545039,Choeroniscus_godmani{T}:6.112672234){T}:10.9120772,((Anoura_latidens{R}:3.779427833,Anoura_geoffroyi{R}:3.779427833){R}:2.83493221,Anoura_caudifer{R}:6.614360043){R}:10.41038939){R}:2.388217391){R}:3.771706573,(Lonchorhina_aurita{R}:8.795556441,Lonchorhina_inusitata{R}:8.795556441){R}:14.38911695){R}:0.8447231842){R}:0.9614243908,(((((Lophostoma_evotis{R}:2.279691562,Lophostoma_silvicolaFG{R}:2.279691562){R}:7.690705428,Lophostoma_brasiliense{R}:9.97039699){R}:7.599298001,(Phyllostomus_elongatus{R}:8.119582824,Phyllostomus_discolor{R}:8.119582824){R}:9.450112167){R}:1.521869862,(Tonatia_saurophila{R}:4.502755885,Tonatia_bidens{R}:4.502755885){R}:14.58880897){R}:3.674523748,(Chrotopterus_auritus{R}:19.15333438,Mimon_cozumelae{R}:19.15333438){R}:3.61275422){R}:2.224732367){R}:2.433669076,((Diaemus_youngi{R}:12.39244932,Desmodus_rotundus{R}:12.39244932){R}:7.835663972,Diphylla_ecaudata{R}:20.22811329){R}:7.196376756){R}:0.8214752215,(((Micronycteris_microtis{R}:9.018674346,Micronycteris_hirsuta{R}:9.018674346){R}:5.324423535,(Micronycteris_minuta{R}:4.882043938,Micronycteris_schmidtorum{R}:4.882043938){R}:9.461053943){R}:8.513447254,Lampronycteris_brachyotis{R}:22.85654514){R}:5.389420131){R}:7.944937657,((((Pteronotus_quadridens{T}:6.670886247,Pteronotus_macleayii{T}:6.670886247){T}:4.04633203,Pteronotus_davyi{T}:10.71721828){T}:3.138003269,Pteronotus_parnellii{T}:13.85522155){T}:18.91999925,Mormoops_megalophylla{T}:32.7752208){T}:3.415682128){R}:5.869690062,Thyroptera_lavali{T}:42.06059299){R}:13.44742223,((((((((Miniopterus_aelleni{R}:1.299615,Miniopterus_petersoni{R}:1.299615){R}:4.270025,Miniopterus_gleni{R}:5.569641){R}:2.003838,Miniopterus_sororculus{R}:7.573479){R}:2.031566,Miniopterus_minor{R}:9.605045){R}:14.889341,(Miniopterus_fuliginosus{R}:17.236618,Miniopterus_schreibersii{R}:17.236618){R}:7.257769){R}:27.640976,Myotis_elegans{T}:52.13536){R}:1.662856,Molossus_sinaloae{T}:53.798217){R}:0.976298,Chilonatalus_micropus{T}:54.774513){R}:0.873379);
Load this file into HyPhy-vision
Results from the first run (Important results are in the .json file):
Test for selection relaxation (K = 0.43) was significant (p = 0.0000, LR = 46.62)
|
|---|
Model (click to enlarge): Alternative (A), General Descriptive (B)
ω distributions under the Alternative model
Test branches are shown in blue and reference branches are shown in red
ω distributions under the Null model
Test branches are shown in blue and reference branches are shown in red
ω distributions under the Partitioned Exploratory model
Test branches are shown in blue and reference branches are shown in red
ω distributions under the Partitioned MG94xREV model
Test branches are shown in blue and reference branches are shown in red
The x-axis of the graphs comes out horribly, but other than that, I'm pleased with the visualization package.