Thinking it doesn't make any sense that BIO_13 precipitation of the warmest month is most important. I also included BIO_18 precipitation of the warmest quarter.
Due to correlation values, you have to make a decision when the going gets rough. I am now going to include BIO_12 [Annual Precipitation], keep 16, get rid of 13 and 14.
R script:
#load packages
library(raster)
library(maptools)
library(dismo)
library(rJava) #also make sure to put maxent.jar into R dismo library folder
#convert to raster
altitude<-raster("/Volumes/LOLOYOHE BA/Merged_layers/alt_seasia9tile.grd")
bio2<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio2_seasia9tile.grd")
bio2<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio2_seasia9tile.grd")
bio5<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio5_seasia9tile.grd")
bio8<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio8_seasia9tile.grd")
bio12<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio12_seasia9tile.grd")
bio15<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio15_seasia9tile.grd")
bio16<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio16_seasia9tile.grd")
bio18<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio18_seasia9tile.grd")
bio19<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio19_seasia9tile.grd")
#stack the layers into one raster and name the layers within the stack
stacked.layers<-stack(altitude, bio2, bio5, bio8, bio12, bio15, bio16, bio18, bio19)
names(stacked.layers)<-c("altitude", "bio2", "bio5", "bio8", "bio12", "bio15", "bio16", "bio18", "bio19")
#remove the original rasters for space
rm(altitude, bio2, bio5, bio8, bio12, bio15, bio16, bio18, bio19)
#read locality points for first species: alcippe peracensis
peracensis.pts<-readShapePoints("/Volumes/LOLOYOHE BA/Vietnam Data/maxent_models/alcippe_peracensis_all.shp")
#run maxent for first species
maxent.peracensis<-maxent(stacked.layers, coordinates(peracensist.pts)[,1:2])
#see graph of important variables
plot(maxent.peracensis)
#see the response curves
response(maxent.peracensis)
#make raster from predictions
r.peracensis<-predict(maxent.peracensis, stacked.layers, progress = "window")
These are my models for six species showing differences between before and after changing the BIOCLIM variables to include. Notice the before is run using the maxent.jar applet rather than with R so I am still figuring out how to get all the same outputs
Alcippe peracensis Mountain fulvetta
Before:
After:
After:
Pellorneum albiventre Spot-throated babbler
Before:
After:
Pomatorhinus ruficollis Streak-breasted scimitar babbler
Before:
Pteruthius
Questions to discuss:
R script:
#load packages
library(raster)
library(maptools)
library(dismo)
library(rJava) #also make sure to put maxent.jar into R dismo library folder
#convert to raster
altitude<-raster("/Volumes/LOLOYOHE BA/Merged_layers/alt_seasia9tile.grd")
bio2<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio2_seasia9tile.grd")
bio2<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio2_seasia9tile.grd")
bio5<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio5_seasia9tile.grd")
bio8<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio8_seasia9tile.grd")
bio12<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio12_seasia9tile.grd")
bio15<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio15_seasia9tile.grd")
bio16<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio16_seasia9tile.grd")
bio18<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio18_seasia9tile.grd")
bio19<-raster("/Volumes/LOLOYOHE BA/Merged_layers/bio19_seasia9tile.grd")
#stack the layers into one raster and name the layers within the stack
stacked.layers<-stack(altitude, bio2, bio5, bio8, bio12, bio15, bio16, bio18, bio19)
names(stacked.layers)<-c("altitude", "bio2", "bio5", "bio8", "bio12", "bio15", "bio16", "bio18", "bio19")
#remove the original rasters for space
rm(altitude, bio2, bio5, bio8, bio12, bio15, bio16, bio18, bio19)
#read locality points for first species: alcippe peracensis
peracensis.pts<-readShapePoints("/Volumes/LOLOYOHE BA/Vietnam Data/maxent_models/alcippe_peracensis_all.shp")
#run maxent for first species
maxent.peracensis<-maxent(stacked.layers, coordinates(peracensist.pts)[,1:2])
#see graph of important variables
plot(maxent.peracensis)
#see the response curves
response(maxent.peracensis)
#make raster from predictions
r.peracensis<-predict(maxent.peracensis, stacked.layers, progress = "window")
These are my models for six species showing differences between before and after changing the BIOCLIM variables to include. Notice the before is run using the maxent.jar applet rather than with R so I am still figuring out how to get all the same outputs
Alcippe peracensis Mountain fulvetta
Before:
| Variable | Percent contribution | Permutation importance |
|---|---|---|
| bio13_seasia9tile | 61.1 | 61.4 |
| bio14_seasia9tile | 18.1 | 4.3 |
| bio15_seasia9tile | 9.4 | 28.4 |
| _alt_seasia9tile | 7.4 | 0.7 |
| _bio2_seasia9tile | 3.3 | 1.3 |
| bio8_seasia9tile | 0.6 | 2 |
| _bio5_seasia9tile | 0.2 | 1.9 |
After:
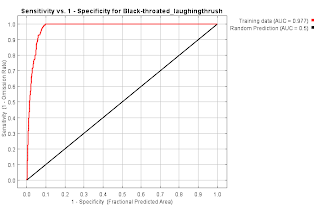
Garrulax chinensis Black-throated laughingthrush
Before:
| Variable | Percent contribution | Permutation importance |
|---|---|---|
| bio13_seasia9tile | 53.7 | 52.6 |
| bio15_seasia9tile | 25.7 | 18.2 |
| bio14_seasia9tile | 6 | 4.3 |
| bio8_seasia9tile | 5.5 | 1 |
| _bio2_seasia9tile | 4 | 1.3 |
| _bio5_seasia9tile | 3.9 | 17.4 |
| _alt_seasia9tile | 1.1 | 5.2 |
After:
Garrulax leucolophus White-crested laughingthrush
Before:
| Variable | Percent contribution | Permutation importance |
|---|---|---|
| bio13_seasia9tile | 58.6 | 29.3 |
| _bio5_seasia9tile | 11.3 | 6.5 |
| _alt_seasia9tile | 9 | 18.8 |
| _bio2_seasia9tile | 7.6 | 13.1 |
| bio14_seasia9tile | 5.5 | 7.1 |
| bio15_seasia9tile | 5.1 | 1.5 |
| bio8_seasia9tile | 2.9 | 23.7 |
Pellorneum albiventre Spot-throated babbler
Before:
| Variable | Percent contribution | Permutation importance |
|---|---|---|
| bio13_seasia9tile | 57.5 | 74.3 |
| _alt_seasia9tile | 17.4 | 1.9 |
| bio15_seasia9tile | 12.9 | 6.5 |
| bio14_seasia9tile | 11.8 | 16.9 |
| _bio2_seasia9tile | 0.2 | 0.5 |
| _bio5_seasia9tile | 0.1 | 0 |
| bio8_seasia9tile | 0 | 0 |
Pomatorhinus ruficollis Streak-breasted scimitar babbler
Before:
| Variable | Percent contribution | Permutation importance |
|---|---|---|
| bio13_seasia9tile | 58.6 | 29.3 |
| _bio5_seasia9tile | 11.3 | 6.5 |
| _alt_seasia9tile | 9 | 18.8 |
| _bio2_seasia9tile | 7.6 | 13.1 |
| bio14_seasia9tile | 5.5 | 7.1 |
| bio15_seasia9tile | 5.1 | 1.5 |
| bio8_seasia9tile | 2.9 | 23.7 |
After:
Pteruthius
Should I even bother including in my analysis anymore? (No longer a babbler)
Questions to discuss:
- Is it okay to rerun models with new set of BIOCLIM variables even if you have already run it once and just notice the first one does not seem biologically true? What if these new variables don't seem right either? Is it okay to keep trying to rerun the models?
- What does it mean when only one variable is giving strong response?
- Precipitation of the Wettest Month (BIO13) was removed as we had thought this didn't make much biological sense due to the way the rainy season works in SE Asia (May/June-Sept/October). We thought it made more sense if Precipitation of the Wettest Quarter were included instead (BIO 16). However, this only seemed to be important for P. ruficollis (as well as altitude). Annual precipitation seemed to be the only important variable (BIO 12) for all other species and nothing else. How can we interpret this?
- How do I make the ROC curve in R?
- I want to validate the model and move forward from this.