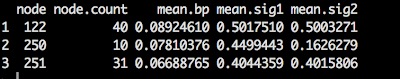
After running GARLI, I wanted to compare
OR137:
OR213: lots of disagreements; redo with 100
OR4: lots of disagreements; redo with 100
OR589: lot
OR6: all agree after 8 runs
OR10: all agree after 8 runs
OR11: tree2 disagrees, 7 others agree, use this as main gene tree
OR51: lots of disagreements; redo with 100
OR52: lots of disagreements; redo with 100
100 is a lot of tree to compare, so I wrote a small script doing so:
setwd("/Volumes/Yango/Hayden_batORs/garli/OR51/trees")
#tell where to open up all of your downloaded files from CIPRES
my.files<-list.files("/Volumes/Yango/Hayden_batORs/garli/OR51/trees/")
#read in all of these trees to a list using read.nexus function
my.trees<-lapply(my.files, read.nexus)
#create a matrix to store all of the tree comparisons
tree.matrix<-matrix(nrow = length(my.trees), ncol = length(my.trees))
for(i in 1:length(my.trees)){ #for every tree in the file
this.tree<-as.phylo(my.trees[[i]]) #look at that tree
#compare the topology to all the other trees
tree.matrix[i,]<-sapply(my.trees, function(my.trees) dist.topo(this.tree,my.trees))
}
tree.matrix
#print out the ones that agree with each other
for(i in 1:length(tree.matrix[1,])){
this.list<-as.numeric(which(tree.matrix[i,]==0))
print(this.list)
}
write.csv(tree.matrix, "tree.matrix.csv")
OR137:
OR213: lots of disagreements; redo with 100
OR4: lots of disagreements; redo with 100
OR589: lot
OR6: all agree after 8 runs
OR10: all agree after 8 runs
OR11: tree2 disagrees, 7 others agree, use this as main gene tree
OR51: lots of disagreements; redo with 100
OR52: lots of disagreements; redo with 100
100 is a lot of tree to compare, so I wrote a small script doing so:
setwd("/Volumes/Yango/Hayden_batORs/garli/OR51/trees")
#tell where to open up all of your downloaded files from CIPRES
my.files<-list.files("/Volumes/Yango/Hayden_batORs/garli/OR51/trees/")
#read in all of these trees to a list using read.nexus function
my.trees<-lapply(my.files, read.nexus)
#create a matrix to store all of the tree comparisons
tree.matrix<-matrix(nrow = length(my.trees), ncol = length(my.trees))
for(i in 1:length(my.trees)){ #for every tree in the file
this.tree<-as.phylo(my.trees[[i]]) #look at that tree
#compare the topology to all the other trees
tree.matrix[i,]<-sapply(my.trees, function(my.trees) dist.topo(this.tree,my.trees))
}
tree.matrix
#print out the ones that agree with each other
for(i in 1:length(tree.matrix[1,])){
this.list<-as.numeric(which(tree.matrix[i,]==0))
print(this.list)
}
write.csv(tree.matrix, "tree.matrix.csv")
For OR51, are things looking better?
[1] 1
[1] 2
[1] 3
[1] 4 84
[1] 5
[1] 6
[1] 7
[1] 8 15 41 79
[1] 9 14
[1] 10 54
[1] 11
[1] 12
[1] 13 73
[1] 9 14
[1] 8 15 41 79
[1] 16
[1] 17 20 75
[1] 18
[1] 19
[1] 17 20 75
[1] 21 51 68
[1] 22 37
[1] 23 50 71
[1] 24
[1] 25
[1] 26
[1] 27
[1] 28 69 88
[1] 29
[1] 30
[1] 31
[1] 32 46
[1] 33
[1] 34 70
[1] 35 53
[1] 36
[1] 22 37
[1] 38 59
[1] 39 47 78
[1] 40
[1] 8 15 41 79
[1] 42
[1] 43
[1] 44
[1] 45
[1] 32 46
[1] 39 47 78
[1] 48
[1] 49
[1] 23 50 71
[1] 21 51 68
[1] 52
[1] 35 53
[1] 10 54
[1] 55
[1] 56
[1] 57
[1] 58
[1] 38 59
[1] 60
[1] 61
[1] 62 86 87
[1] 63
[1] 64
[1] 65
[1] 66 95
[1] 67
[1] 21 51 68
[1] 28 69 88
[1] 34 70
[1] 23 50 71
[1] 72
[1] 13 73
[1] 74
[1] 17 20 75
[1] 76
[1] 77
[1] 39 47 78
[1] 8 15 41 79
[1] 80
[1] 81
[1] 82
[1] 83
[1] 4 84
[1] 85
[1] 62 86 87
[1] 62 86 87
[1] 28 69 88
[1] 89
[1] 90
[1] 91
[1] 92
[1] 93
[1] 94
[1] 66 95
[1] 96
[1] 97
[1] 98
Womp, womp...not really--only a maximum of 4 trees out of 100 are agreeing.
OR4s looking much better:
[1] 1 22 28 42 75
[1] 2 99
[1] 3 14 18 83 89
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 5
[1] 6
[1] 7 15 98
[1] 8
[1] 9 19 38 80
[1] 10 50
[1] 11 67
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 13 86
[1] 3 14 18 83 89
[1] 7 15 98
[1] 16
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 3 14 18 83 89
[1] 9 19 38 80
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 21 31 44 62 84
[1] 1 22 28 42 75
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 25
[1] 26
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 1 22 28 42 75
[1] 29
[1] 30 33
[1] 21 31 44 62 84
[1] 32 37 61 68 81
[1] 30 33
[1] 34 100
[1] 35
[1] 36
[1] 32 37 61 68 81
[1] 9 19 38 80
[1] 39 71
[1] 40 49
[1] 41
[1] 1 22 28 42 75
[1] 43
[1] 21 31 44 62 84
[1] 45
[1] 46
[1] 47
[1] 48
[1] 40 49
[1] 10 50
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 52
[1] 53
[1] 54
[1] 55
[1] 56 90
[1] 57
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 60
[1] 32 37 61 68 81
[1] 21 31 44 62 84
[1] 63
[1] 64
[1] 65
[1] 66
[1] 11 67
[1] 32 37 61 68 81
[1] 69
[1] 70
[1] 39 71
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 73
[1] 74
[1] 1 22 28 42 75
[1] 76
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 78
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 9 19 38 80
[1] 32 37 61 68 81
[1] 82
[1] 3 14 18 83 89
[1] 21 31 44 62 84
[1] 85
[1] 13 86
[1] 87
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 3 14 18 83 89
[1] 56 90
[1] 91
[1] 92
[1] 93
[1] 94
[1] 95
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 97
[1] 7 15 98
[1] 2 99
[1] 34 100
OR4s looking much better:
[1] 1 22 28 42 75
[1] 2 99
[1] 3 14 18 83 89
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 5
[1] 6
[1] 7 15 98
[1] 8
[1] 9 19 38 80
[1] 10 50
[1] 11 67
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 13 86
[1] 3 14 18 83 89
[1] 7 15 98
[1] 16
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 3 14 18 83 89
[1] 9 19 38 80
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 21 31 44 62 84
[1] 1 22 28 42 75
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 25
[1] 26
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 1 22 28 42 75
[1] 29
[1] 30 33
[1] 21 31 44 62 84
[1] 32 37 61 68 81
[1] 30 33
[1] 34 100
[1] 35
[1] 36
[1] 32 37 61 68 81
[1] 9 19 38 80
[1] 39 71
[1] 40 49
[1] 41
[1] 1 22 28 42 75
[1] 43
[1] 21 31 44 62 84
[1] 45
[1] 46
[1] 47
[1] 48
[1] 40 49
[1] 10 50
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 52
[1] 53
[1] 54
[1] 55
[1] 56 90
[1] 57
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 60
[1] 32 37 61 68 81
[1] 21 31 44 62 84
[1] 63
[1] 64
[1] 65
[1] 66
[1] 11 67
[1] 32 37 61 68 81
[1] 69
[1] 70
[1] 39 71
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 73
[1] 74
[1] 1 22 28 42 75
[1] 76
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 78
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 9 19 38 80
[1] 32 37 61 68 81
[1] 82
[1] 3 14 18 83 89
[1] 21 31 44 62 84
[1] 85
[1] 13 86
[1] 87
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 3 14 18 83 89
[1] 56 90
[1] 91
[1] 92
[1] 93
[1] 94
[1] 95
[1] 4 12 17 20 23 24 27 51 58 59 72 77 79 88 96
[1] 97
[1] 7 15 98
[1] 2 99
[1] 34 100